6. Erdos-Renyi random networks#
import random
import networkx as nx
import numpy as np
import seaborn as sb
import matplotlib.pyplot as plt
sb.set_theme(style="ticks", context="notebook")
random.random()
0.6529223135709693
6.1. Write a random graph generator#
def random_graph(N, p):
G = nx.Graph()
nodes = range(N)
G.add_nodes(N)
for i in nodes:
for j in nodes[:i]:
if random.random <= p:
G.add_edge((i, j))
return G
def random_graph(N, p):
G = nx.Graph()
nodes = range(N)
G.add_nodes_from(nodes)
edges = []
for i in nodes:
for j in nodes[:i]:
if random.random() < p:
edges.append([i, j])
G.add_edges_from(edges)
return G
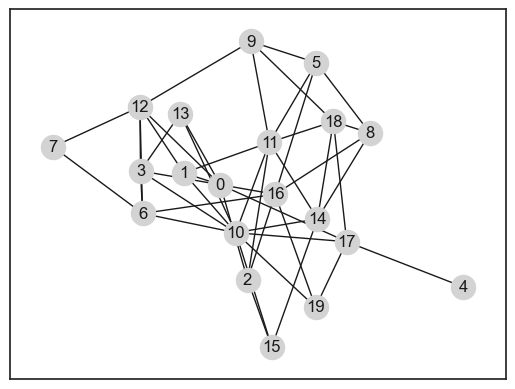
G = random_graph(N=10, p=0.1)
print(G)
fig, ax = plt.subplots(figsize=(3, 3))
nx.draw(G, node_size=15)
Graph with 10 nodes and 3 edges

N = 100
p = 0.8 / N
G = random_graph(N, p)
np.log(N)
4.605170185988092
params = {
"node_size": 10,
"with_labels": False,
"edge_color": "silver",
"node_color": "b",
}
nx.draw_networkx(G, **params)
sb.despine(bottom=True, left=True)
plt.show()

6.2. Analyse characteristics#
N = 1000
p = 5 / N
G = nx.erdos_renyi_graph(N, p, seed=1)
params = {
"node_size": 10,
"with_labels": False,
"edge_color": "silver",
"node_color": "b",
}
pos = nx.spring_layout(G)
nx.draw_networkx(G, pos=pos, **params)
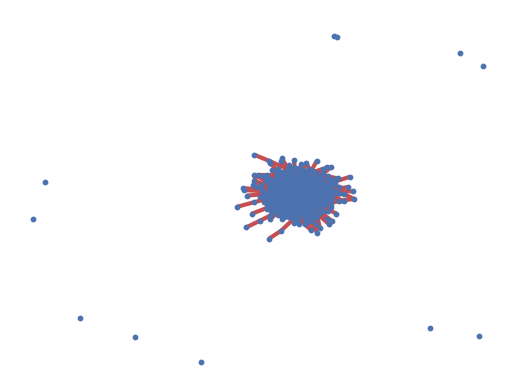
# identify largest connected component
Gcc = sorted(nx.connected_components(G), key=len, reverse=True)
G0 = G.subgraph(Gcc[0])
# highlight largest connected component
nx.draw_networkx_edges(G0, pos=pos, width=3.0, edge_color="r")
# draw other connected components
for Gi in Gcc[1:]:
if len(Gi) > 1:
nx.draw_networkx_edges(
G.subgraph(Gi), pos, alpha=0.4, width=3.0, edge_color="r"
)
sb.despine(bottom=True, left=True)
/var/folders/wm/5gv37br900l73y63tjf8sr1r0000gn/T/ipykernel_7931/3661388696.py:21: DeprecationWarning: `alltrue` is deprecated as of NumPy 1.25.0, and will be removed in NumPy 2.0. Please use `all` instead.
nx.draw_networkx_edges(
print(f"Connected: {nx.is_connected(G)}")
print(f"# connected components: {len(list(nx.connected_components(G)))}")
print()
print(f"Size of largest connected component: {len(G0)}")
print(f"Prop. of nodes in it: {len(G0) / N:.2f}")
print()
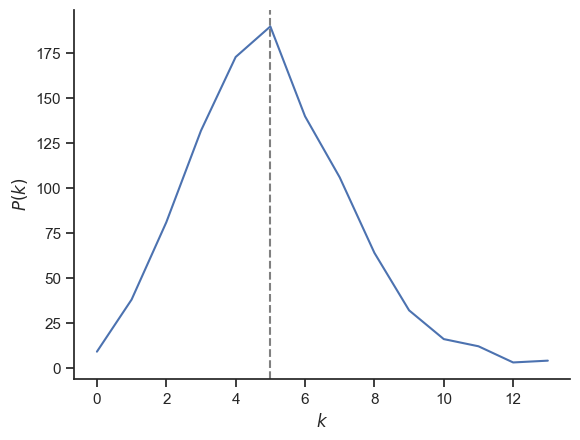
degree_sequence = sorted((d for n, d in G.degree()), reverse=True)
print(f"Average degree: {np.average(degree_sequence)}")
print(f"Clustering coefficient: {nx.average_clustering(G)}")
Connected: False
# connected components: 11
Size of largest connected component: 989
Prop. of nodes in it: 0.99
Average degree: 5.0
Clustering coefficient: 0.005966788766788765
deg, counts = np.unique(degree_sequence, return_counts=True)
fig, ax = plt.subplots()
ax.plot(deg, counts)
ax.axvline(np.average(degree_sequence), ls="--", c="grey", zorder=-1)
ax.set_xlabel("$k$")
ax.set_ylabel("$P(k)$")
#plt.yscale("log")
#plt.xscale("log")
sb.despine()
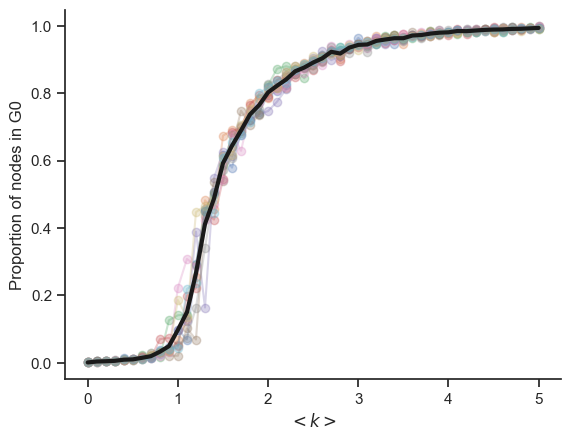
6.3. Vary the degree k#
N = 1000
ks = np.
N = 1000
ks = np.arange(0, 5.1, 0.1)
ps = ks / (N - 1)
n_reps = 10
props_arr = np.zeros((len(ps), n_reps))
for i, p in enumerate(ps):
for rep in range(n_reps):
G = nx.erdos_renyi_graph(N, p)
Gcc = sorted(nx.connected_components(G), key=len, reverse=True)
G0 = G.subgraph(Gcc[0])
prop = len(G0) / N
props_arr[i, rep] = prop
fig, ax = plt.subplots()
ax.plot(ks, props_arr[:, :], "-o", alpha=0.3)
ax.plot(ks, np.average(props_arr, axis=1), "k-", lw=3)
ax.set_ylabel("Proportion of nodes in G0")
ax.set_xlabel(r"$< k >$")
sb.despine()
6.4. Configuration model#
N = 20
p = 6 / N
G = random_graph(N, p)
degree_sequence = [d for n, d in G.degree()]
degree_sequence
[7, 4, 4, 7, 1, 4, 5, 2, 6, 5, 9, 7, 9, 3, 5, 5, 6, 5, 5, 3]
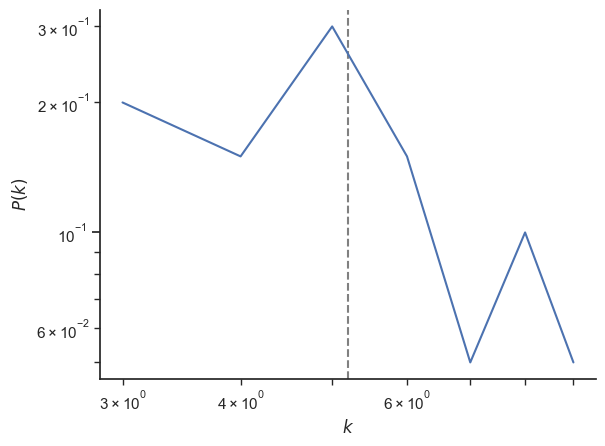
deg, counts = np.unique(degree_sequence, return_counts=True)
fig, ax = plt.subplots()
ax.plot(deg, counts / N)
ax.axvline(np.average(degree_sequence), ls="--", c="grey", zorder=-1)
ax.set_xlabel("$k$")
ax.set_ylabel("$P(k)$")
plt.yscale("log")
plt.xscale("log")
sb.despine()
G_conf = nx.configuration_model(degree_sequence)
print(G_conf)
MultiGraph with 20 nodes and 51 edges
G_conf = nx.Graph(G_conf)
print(G_conf)
Graph with 20 nodes and 48 edges
degree_sequence_conf = [d for n, d in G_conf.degree()]
print(G)
Graph with 20 nodes and 51 edges
print(G_conf)
Graph with 20 nodes and 48 edges
deg_conf, counts_conf = np.unique(degree_sequence_conf, return_counts=True)
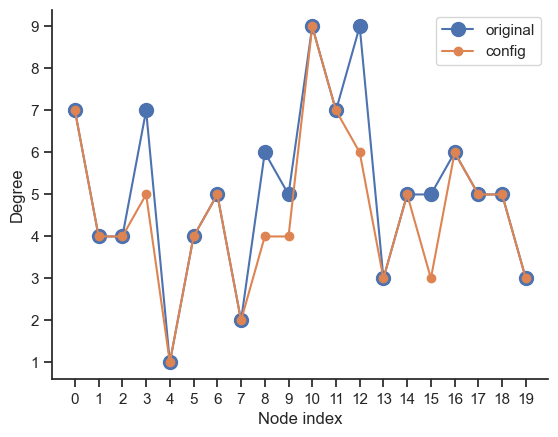
fig, ax = plt.subplots()
ax.plot(degree_sequence, "o-", ms=10, label="original")
ax.plot(degree_sequence_conf, "o-", label="config")
ax.set_xlabel("Node index")
ax.set_ylabel("Degree")
# plt.yscale("log")
# plt.xscale("log")
ax.set_xticks(range(len(G)))
ax.legend()
sb.despine()
print(G)
print(G_conf)
Graph with 20 nodes and 51 edges
Graph with 20 nodes and 48 edges
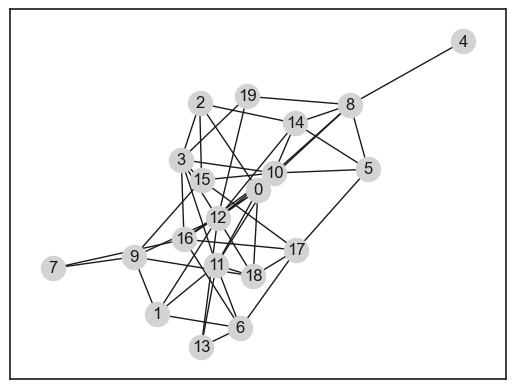
pos = nx.spring_layout(G)
pos = nx.spring_layout(G, seed=1)
nx.draw_networkx(G, pos=pos, node_color="lightgrey")
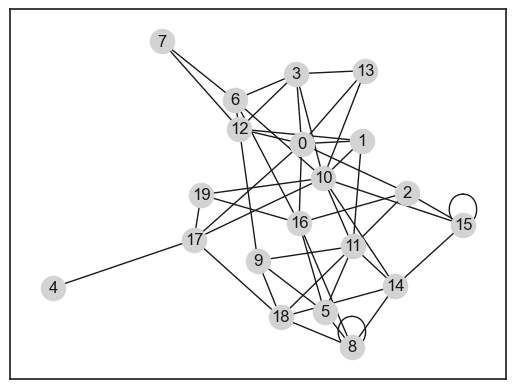
nx.draw_networkx(G_conf, node_color="lightgrey")
list(nx.selfloop_edges(G_conf))
[(8, 8), (15, 15)]
print(G_conf)
Graph with 20 nodes and 48 edges
edges_to_remove = list(nx.selfloop_edges(G_conf))
edges_to_remove
[(8, 8), (15, 15)]
G_conf.remove_edges_from(edges_to_remove)
print(G_conf)
Graph with 20 nodes and 46 edges
edges_to_remove
[(12, 12)]
nx.draw_networkx(G_conf, node_color="lightgrey")